Connections: Wide Field Virtual Array Imaging - Innovation at the Microscopic Scale
The Foundation of Disruption From A Scientific Imaging Perspective
Considering scientific imaging, the common application of microscopes for research can span a variety of imaging modalities to examine things ranging from whole organism to atomic level. For example, a light microscope, at best, can resolve an object that is 0.5 microns (5x10-7 meters) in width, approximately the size of a bacterium. Anything smaller (cellular structures, individual proteins, individual molecules) cannot be seen. This limitation is due to the properties of visible light and cannot be breached by improving the mechanics of the light microscope.
Electron microscopy, by its design, employs electrons at a wavelength much smaller than visible light thus enabling structures smaller than a bacterium to be made visible. Numerous software-based techniques are being developed to further push the resolving limits of light microscopy to achieve 'near molecular resolution'. But whether you agree or not, electron microscopy (EM) still reigns supreme for ultimate structural examination of cellular proteins and the cellular microenvironment by virtue of the imaging source wavelength.
However, like traditional light microscopy with fixed objective lenses, pre-defined magnification settings on an electron microscope result in increasing narrowing fields of view as magnification levels are increased. Many researchers not only want detailed structures, they also want to see a larger view, i.e., 'the bigger picture'. Currently, technology exists that enable overcoming several limitations of fixed focal length microscopic observation with a number of unique advantages. One modern application of technology to address and disrupt significant shortcomings to routine light microscopy is Digital Pathology. Likewise, traditional electron microscopic imaging is ripe for disruption and long overdue for change!
Biological EM: A Traditional Workflow
Invented in 1931 and developed for commercial use in 1938, the general workflow for the preparation and viewing of biological samples in a transmission electron microscope has been well-established. Areas of interest are identified and image capture is conducted at low magnifications in order to present general cellular architecture containing low resolution but having a wide field of view (FOV). Higher magnifications are used to focus on areas of interest which contain high resolution data but have a narrow FOV.Historically, photographic films were used for image capture. These film negatives were processed and then printed via photographic darkroom techniques. Now, few labs still employ film methods and most have converted to digital imaging. However, the classical, and time-consuming workflow of capturing individual images continues and results in a cumbersome management of image files well into the hundreds for any given investigation. In addition to the mechanics of image capture and storage, the image file naming system must typically comply with a standardized nomenclature for ease of identification at a later time point, a process which is also cumbersome and time-consuming. The images are usually reviewed on an individual basis and select ones chosen for presentation purposes as 'representative' which introduces bias into the scientific process. While being sufficient for purpose (aka 'easy'), the resulting collection of image files are static, non-interactive, and any individual image file is disconnected from the others (see Figure 1A).
Moving from Static to Dynamic: Wide Field Imaging Array Methodology
-Frank G.A. Fass
As Dr. Fass discovered, the answer to the dilemma of context and detail is wide-field imaging technology! This elegant and highly efficient method produces not a collection of unconnected single images, but a single "image data set" comprised of an array of tens to tens-of-thousands of high resolution images each with a narrow FOV, which are then stitched together 'on-the-fly' in a tiled format and presented to the viewer as a single virtual image. A combination of toolbar menu items and computer mouse buttons allow for panning and zooming. The nature of these datasets provide context and connection, enabling the end-user to easily transition from low-resolution overview (at the 'top' of the image pyramid) to a detailed examination of specific features (at the 'bottom' of the image pyramid) (see Figure 1B).
Similar to Google Earth, a multitude of high resolution, albeit small and individual FOV image tiles are delivered to the end-user as a single virtual image which provides both overall context and structural detail, all in one highly manageable package.
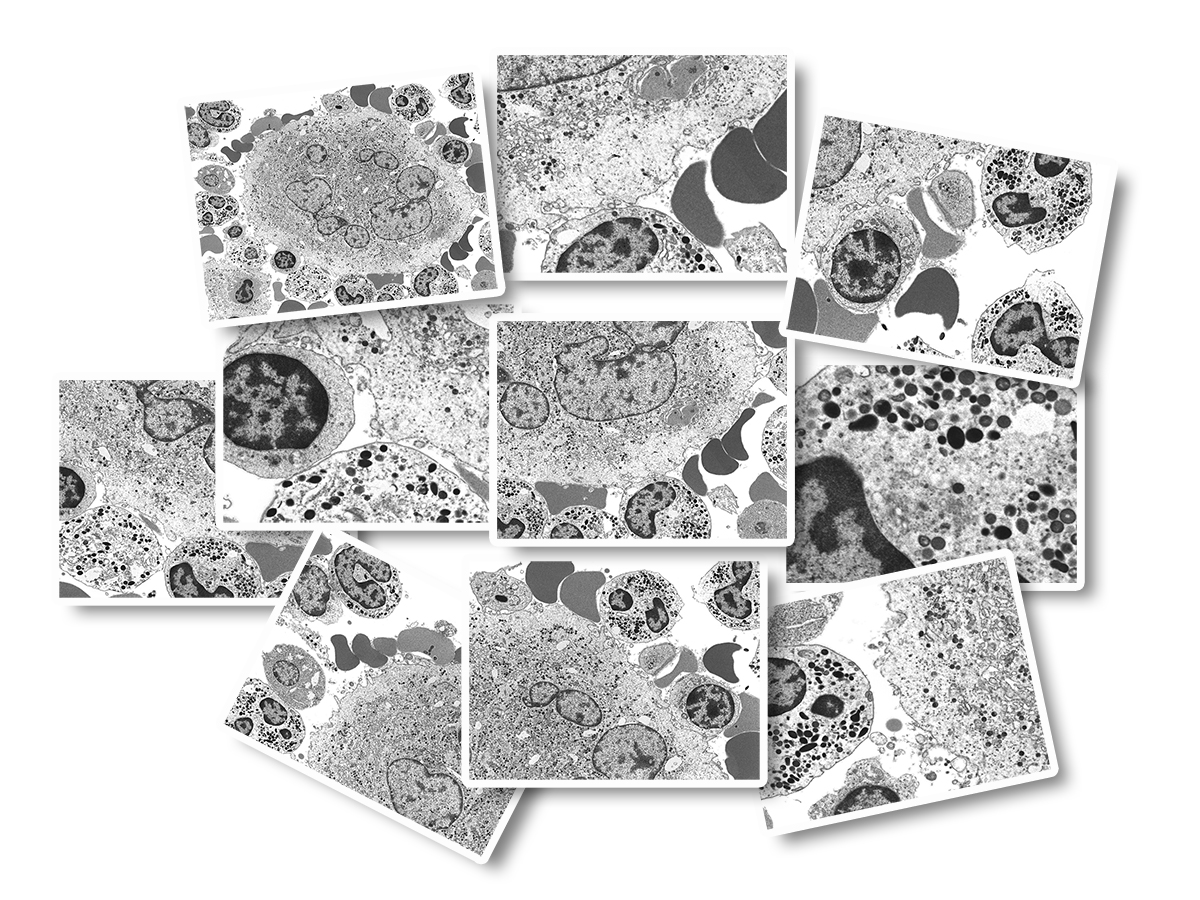
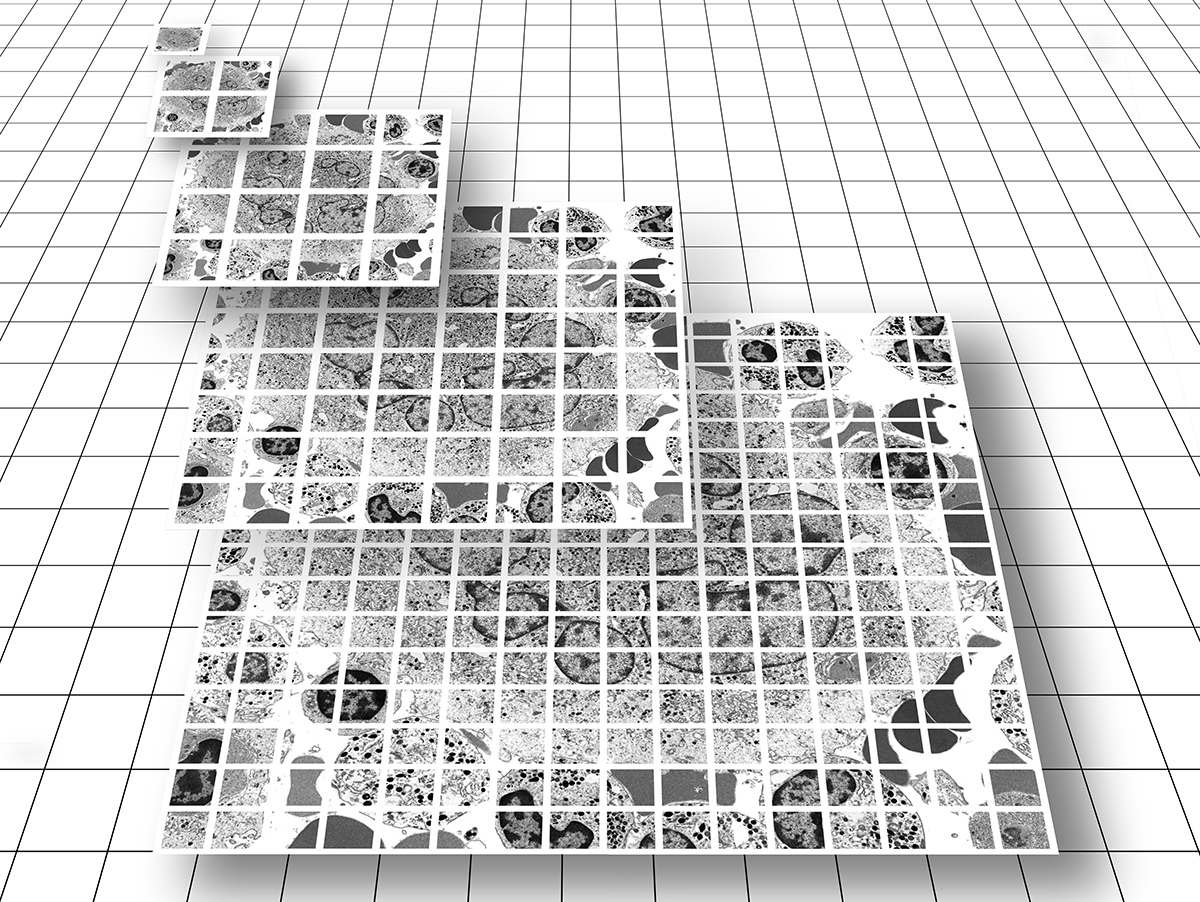
This innovative imaging technology solves a number of problematic issues associated with traditional EM work processes including:
- Maintenance of one dataset file folder. Reduction in time and effort for organizing and maintaining tens or thousands of individual files and associated file names
- Browser-based viewing. No complicated instrumentation controls for inexperienced users means an improved end-user viewing experience
- High Resolution Wide FOV. Combining 'the bigger picture' with high resolvable detail, all within one complete package, enables improved image context and assessment
- Reduction in bias. Presentation of high resolution, wide field data rather than traditional 'single-shot' images promotes scientific transparency by enabling a view beyond the artificial boundaries of regions subjectively designated as 'representative' by the end-user/investigator/author.
- Reallocation of colleague time. Enables focus on higher-value work due to automation of high-resolution array imaging
View examples of zoomable array imaging in my Zoomable Digital Art Gallery!